Protein-ligand Docking tutorial using BioExcel Building Blocks (biobb)
– PDBe REST-API Version –
This tutorial aims to illustrate the process of protein-ligand docking, step by step, using the BioExcel Building Blocks library (biobb). The particular example used is the Mitogen-activated protein kinase 14 (p38-α) protein (PDB code 3LFA, https://doi.org/10.2210/pdb3LFA/pdb), a well-known Protein Kinase enzyme, in complex with the FDA-approved Dasatinib, (PDB Ligand code 1N1, DrugBank Ligand Code DB01254), a small molecule kinase inhibitor used for the treatment of lymphoblastic or chronic myeloid leukemia with resistance or intolerance to prior therapy.
The tutorial will guide you through the process of identifying the active site cavity (pocket) and the final prediction of the protein-ligand complex.
Please note that docking algorithms, and in particular, AutoDock Vina program used in this tutorial, are non-deterministic. That means that results obtained when running the workflow could be diferent from the ones we obtained during the writing of this tutorial (see AutoDock Vina manual). We invite you to try the docking process several times to verify this behaviour.
Settings
Biobb modules used
biobb_io: Tools to fetch biomolecular data from public databases.
biobb_structure_utils: Tools to modify or extract information from a PDB structure file.
biobb_chemistry: Tools to perform chemoinformatics processes.
biobb_vs: Tools to perform virtual screening studies.
Auxiliar libraries used
Conda Installation and Launch
git clone https://github.com/bioexcel/biobb_wf_virtual-screening.git
cd biobb_wf_virtual-screening
conda env create -f conda_env/environment.yml
conda activate biobb_VS_tutorial
jupyter-notebook biobb_wf_virtual-screening/notebooks/ebi_api/wf_vs_ebi_api.ipynb
Pipeline steps

Input parameters
Input parameters needed:
pdb_code: PDB code of the experimental complex structure (if exists).
In this particular example, the p38α structure in complex with the Dasatinib drug was experimentally solved and deposited in the PDB database under the 3LFA PDB code, https://doi.org/10.2210/pdb3LFA/pdb. The protein structure from this PDB file will be used as a target protein for the docking process, after stripping the small molecule. An APO structure, or any other structure from the p38α cluster 100 (sharing a 100% of sequence similarity with the p38α structure) could also be used as a target protein. This structure of the protein-ligand complex will be also used in the last step of the tutorial to check how close the resulting docking pose is from the known experimental structure.
ligandCode: Ligand PDB code (3-letter code) for the small molecule (e.g. 1N1, DrugBank Ligand Code DB01254).
In this particular example, the small molecule chosen for the tutorial is the FDA-approved drug Dasatinib (PDB Code 1N1, DrugBank Ligand Code DB01254), a tyrosine kinase inhibitor, used in lymphoblastic or chronic myeloid leukemia.
import nglview
import ipywidgets
pdb_code = "3LFA" # P38 + Dasatinib
ligand_code = "1N1" # Dasatinib
Fetching PDB structure
Downloading PDB structure with the protein molecule from the PDBe database.
Alternatively, a PDB file can be used as starting structure.
Building Blocks used:
Pdb from biobb_io.api.pdb
from biobb_io.api.pdb import pdb
download_pdb = "download.pdb"
prop = {
"pdb_code": pdb_code,
"filter": ["ATOM", "HETATM"]
}
pdb(output_pdb_path=download_pdb,
properties=prop)
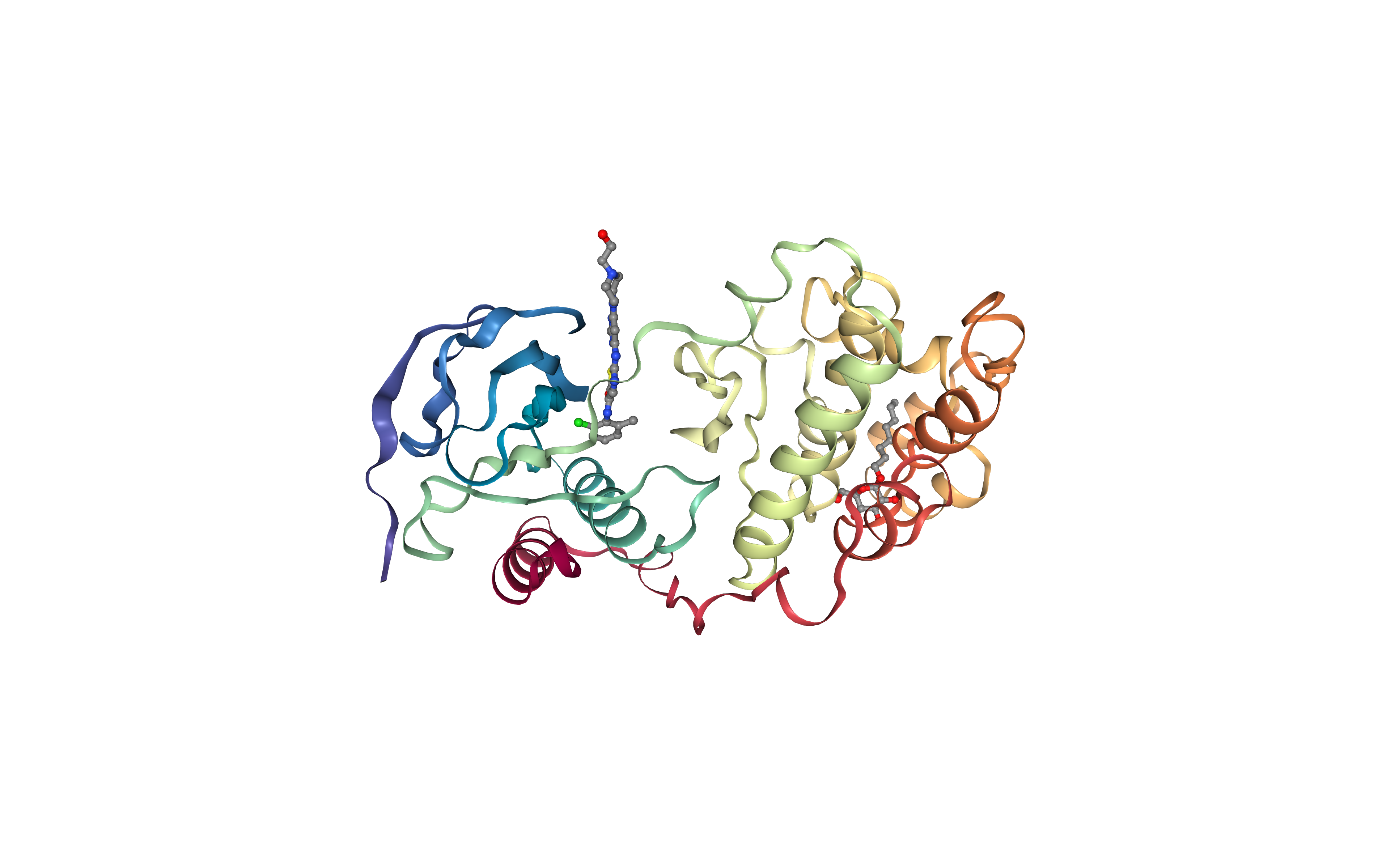
Visualizing 3D structure
Visualizing the downloaded/given PDB structure using NGL.
Note (and try to identify) the Dasatinib small molecule (1N1) and the detergent (β-octyl glucoside) (BOG) used in the experimental reservoir solution to obtain the crystal.
view = nglview.show_structure_file(download_pdb, default=True)
view.center()
view._remote_call('setSize', target='Widget', args=['','600px'])
view
Extract Protein Structure
Extract protein structure from the downloaded PDB file. Removing any extra molecule (ligands, ions, water molecules).
The protein structure will be used as a target in the protein-ligand docking process.
Building Blocks used:
extract_molecule from biobb_structure_utils.utils.extract_molecule
from biobb_structure_utils.utils.extract_molecule import extract_molecule
pdb_protein = "pdb_protein.pdb"
extract_molecule(input_structure_path=download_pdb,
output_molecule_path = pdb_protein)
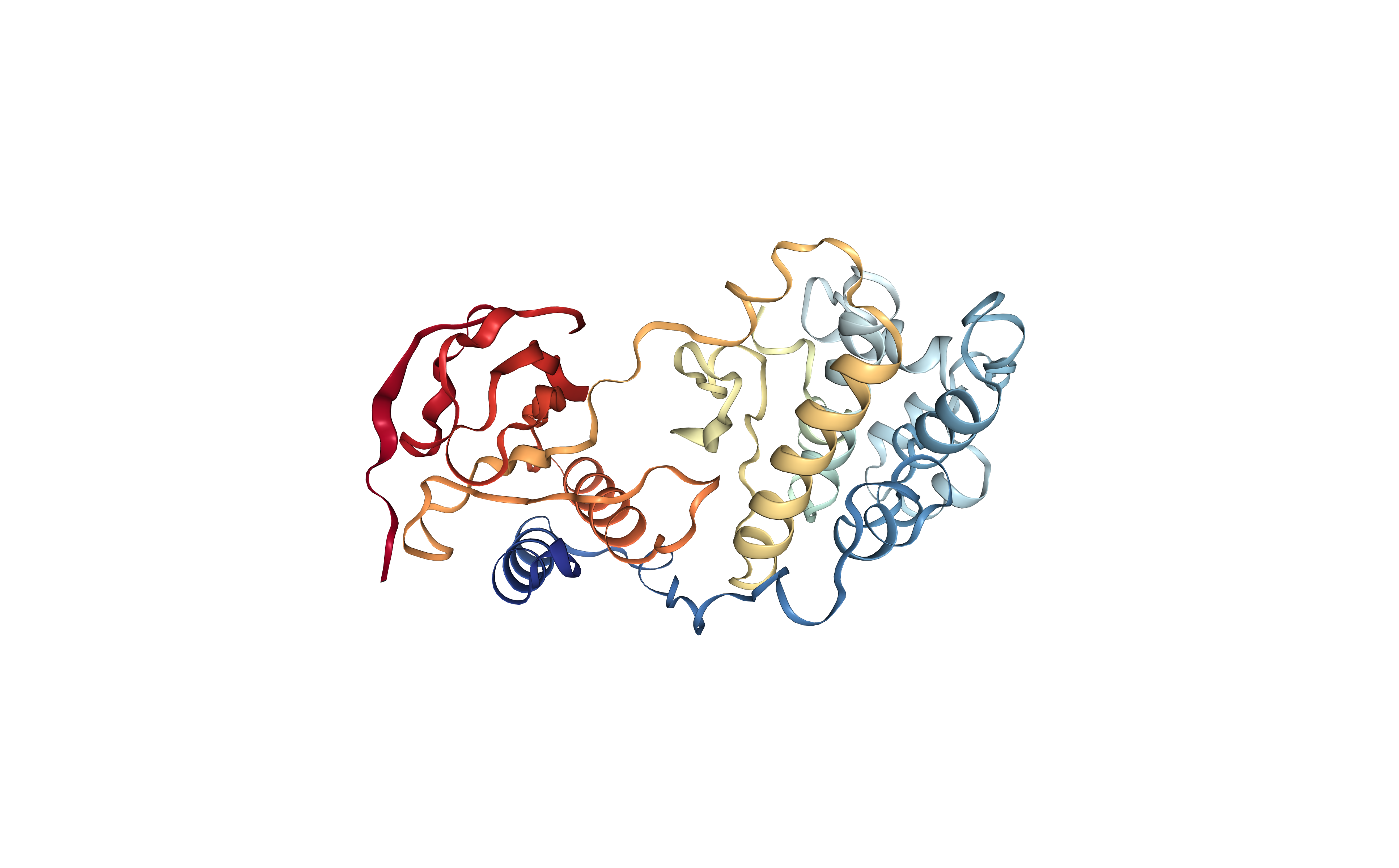
Visualizing 3D structure
Visualizing the downloaded/given PDB structure using NGL.
Note that the small molecules included in the original structure are now gone. The new structure only contains the protein molecule, which will be used as a target for the protein-ligand docking.
view = nglview.show_structure_file(pdb_protein, default=False)
view.add_representation(repr_type='cartoon',
selection='not het',
colorScheme = 'atomindex')
view.center()
view._remote_call('setSize', target='Widget', args=['','600px'])
view
Computing Protein Cavities (PDBe REST-API)
Identifying the protein cavities (pockets) using the PDBe REST-API.
These cavities will be then used in the docking procedure to try to find the best region of the protein surface where the small molecule can bind.
Although in this particular example we already know the binding site region, as we started from a protein-ligand complex structure where the ligand was located in the same binding site as Dasatinib is binding, the PDBe REST-API can be used to automatically identify and extract the possible binding sites of our target protein from the PDB file annotations. This REST-API endpoint provides details on binding sites from the PDB files (or mmcif) information, such as ligands, residues in the site or description of the site.
Building Blocks used:
api_binding_site from biobb_io.api.api_binding_site
from biobb_io.api.api_binding_site import api_binding_site
residues_json = "residues.json"
prop = {
"pdb_code": pdb_code
}
api_binding_site(output_json_path=residues_json,
properties=prop)
Checking binding site output (json)
Checking the PDBe REST-API output from the json file. Every pocket has a separated entry in the json output, with information such as: residues forming the cavity, details of the cavity, and evidence used to detect the cavity.
import json
with open(residues_json) as json_file:
data = json.load(json_file)
print(json.dumps(data, indent=4))
Select binding site (cavity)
Select a specific binding site (cavity) from the obtained list to be used in the docking procedure.
If the PDBe REST-API has successfully identified the correct binding site, which we know from the original protein-ligand structure, it just needs to be selected. In this particular example, the cavity we are interested in is the binding site number 1.
Choose a binding site from the DropDown list:
bindingSites = {}
bsites = []
for i, item in enumerate(data[pdb_code.lower()]):
bindingSites[i] = []
bsites.append(('binding_site' + str(i), i))
for res in item['site_residues']:
bindingSites[i].append(res['author_residue_number'])
# print('Residue id\'s for binding site %d: %s' % (i, ', '.join(str(v) for v in bindingSites[i])))
mdsel = ipywidgets.Dropdown(
options=bsites,
description='Binding Site:',
disabled=False,
)
display(mdsel)
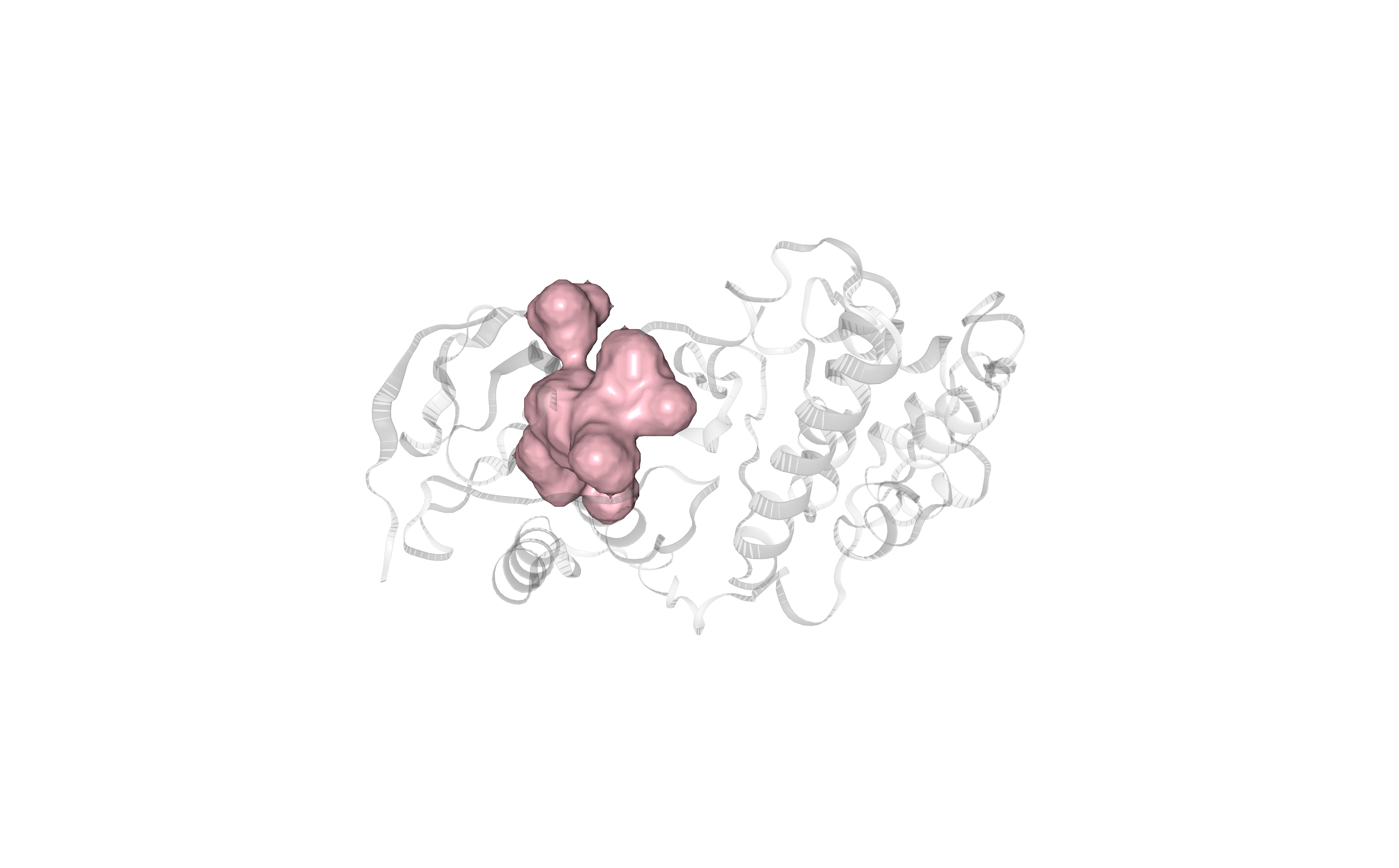
Visualizing selected binding site (cavity)
Visualizing the selected binding site (cavity) using NGL viewer.
Protein residues forming the cavity are represented in pink-colored surface.
view = nglview.show_structure_file(download_pdb, default=False)
view.add_representation(repr_type='cartoon',
selection='not het',
opacity=.2,
color='#cccccc')
view.add_representation(repr_type='surface',
selection=', '.join(str(v) for v in bindingSites[mdsel.value]),
color='pink',
lowResolution= True,
# 0: low resolution
smooth=1 )
view.center()
view._remote_call('setSize', target='Widget', args=['','600px'])
view
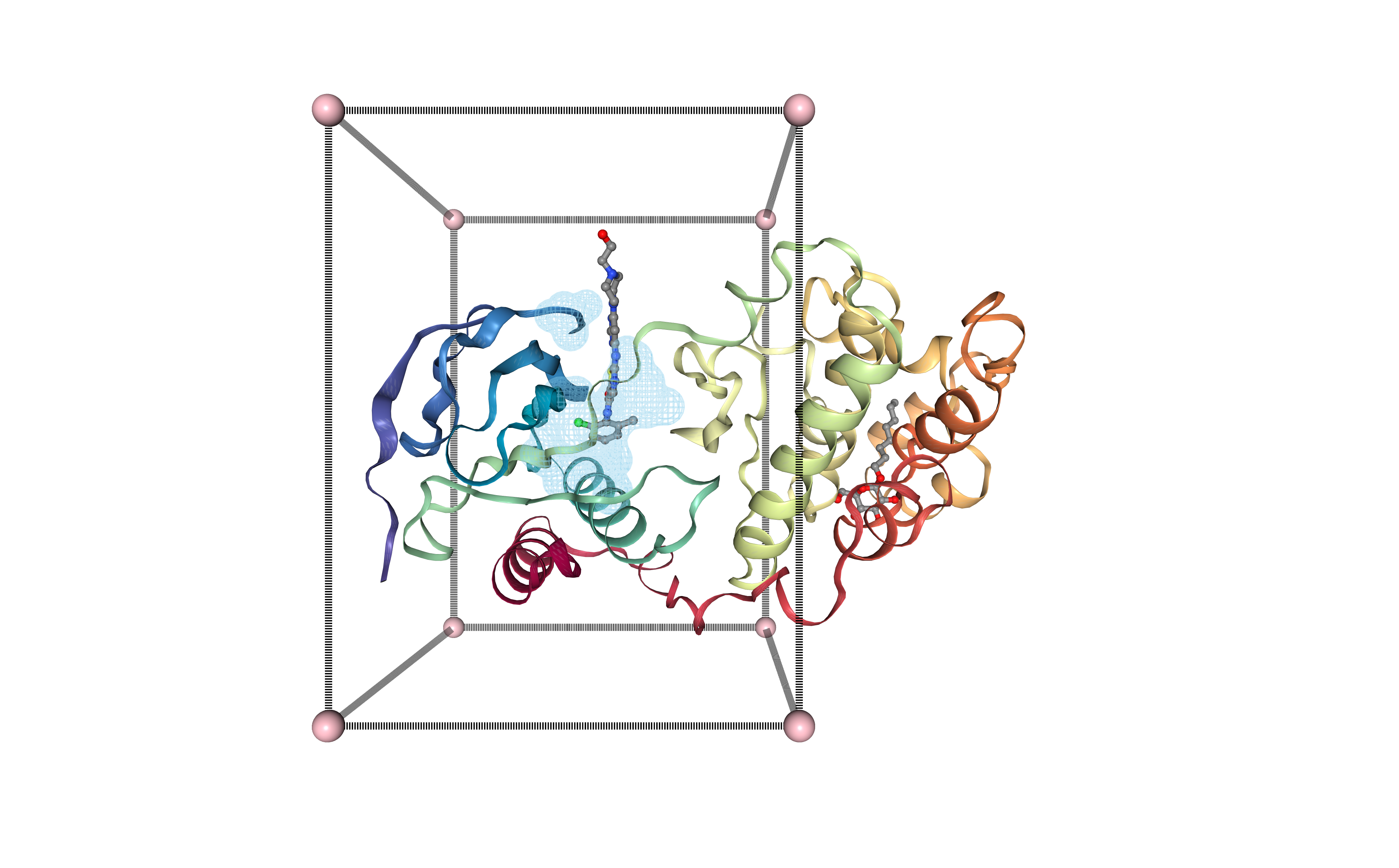
Generating Cavity Box
Generating a box surrounding the selected protein cavity (pocket), to be used in the docking procedure. The box is defining the region on the surface of the protein target where the docking program should explore a possible ligand dock.
An offset of 12 Angstroms is used to generate a big enough box to fit the small molecule and its possible rotations.
Building Blocks used:
box_residues from biobb_vs.utils.box_residues
from biobb_vs.utils.box_residues import box_residues
output_box = "box.pdb"
prop = {
"resid_list": bindingSites[mdsel.value],
"offset": 12,
"box_coordinates": True
}
box_residues(#input_pdb_path = pdb_single_chain,
input_pdb_path = download_pdb,
output_pdb_path = output_box,
properties=prop)
Visualizing binding site box in 3D structure
Visualizing the protein structure, the selected cavity, and the generated box, all together using NGL viewer. Using the original structure with the small ligand inside (Dasatinib, 1N1, DrugBank Ligand Code DB01254), to check that the selected cavity is placed in the same region as the original ligand.
view = nglview.NGLWidget()
s = view.add_component(download_pdb)
b = view.add_component(output_box)
atomPair = [
[ "9999:Z.ZN1", "9999:Z.ZN2" ],
[ "9999:Z.ZN2", "9999:Z.ZN4" ],
[ "9999:Z.ZN4", "9999:Z.ZN3" ],
[ "9999:Z.ZN3", "9999:Z.ZN1" ],
[ "9999:Z.ZN5", "9999:Z.ZN6" ],
[ "9999:Z.ZN6", "9999:Z.ZN8" ],
[ "9999:Z.ZN8", "9999:Z.ZN7" ],
[ "9999:Z.ZN7", "9999:Z.ZN5" ],
[ "9999:Z.ZN1", "9999:Z.ZN5" ],
[ "9999:Z.ZN2", "9999:Z.ZN6" ],
[ "9999:Z.ZN3", "9999:Z.ZN7" ],
[ "9999:Z.ZN4", "9999:Z.ZN8" ]
]
# structure
s.add_representation(repr_type='cartoon',
selection='not het',
color='#cccccc',
opacity=.2)
# ligands box
b.add_representation(repr_type='ball+stick',
selection='9999',
color='pink',
aspectRatio = 8)
# lines box
b.add_representation(repr_type='distance',
atomPair= atomPair,
labelVisible=False,
color= 'black')
# residues
s.add_representation(repr_type='surface',
selection=', '.join(str(v) for v in bindingSites[mdsel.value]),
color='skyblue',
lowResolution= True,
# 0: low resolution
smooth=1,
surfaceType= 'av',
contour=True,
opacity=0.4,
useWorker= True,
wrap= True)
view.center()
view._remote_call('setSize', target='Widget', args=['','600px'])
view
Downloading Small Molecule
Downloading the desired small molecule to be used in the docking procedure.
In this particular example, the small molecule of interest is the FDA-approved drug Dasatinib, with PDB code 1N1.
Building Blocks used:
ideal_sdf from biobb_io.api.ideal_sdf
from biobb_io.api.ideal_sdf import ideal_sdf
sdf_ideal = "ideal.sdf"
prop = {
"ligand_code": ligand_code
}
ideal_sdf(output_sdf_path=sdf_ideal,
properties=prop)
Converting Small Molecule
Converting the desired small molecule to be used in the docking procedure, from SDF format to PDB format using the OpenBabel chemoinformatics tool.
Building Blocks used:
babel_convert from biobb_chemistry.babelm.babel_convert
from biobb_chemistry.babelm.babel_convert import babel_convert
ligand = "ligand.pdb"
prop = {
"input_format": "sdf",
"output_format": "pdb",
"binary_path": "obabel"
}
babel_convert(input_path = sdf_ideal,
output_path = ligand,
properties=prop)
Preparing Small Molecule (ligand) for Docking
Preparing the small molecule structure for the docking procedure. Converting the PDB file to a PDBQT file format (AutoDock PDBQT: Protein Data Bank, with Partial Charges (Q), & Atom Types (T)), needed by AutoDock Vina.
The process adds partial charges and atom types to every atom. Besides, the ligand flexibility is also defined in the information contained in the file. The concept of “torsion tree” is used to represent the rigid and rotatable pieces of the ligand. A rigid piece (”root”) is defined, with zero or more rotatable pieces (”branches”), hanging from the root, and defining the rotatable bonds.
More info about PDBQT file format can be found in the AutoDock FAQ pages.
Building Blocks used:
babel_convert from biobb_chemistry.babelm.babel_convert
from biobb_chemistry.babelm.babel_convert import babel_convert
prep_ligand = "prep_ligand.pdbqt"
prop = {
"input_format": "pdb",
"output_format": "pdbqt",
"binary_path": "obabel"
}
babel_convert(input_path = ligand,
output_path = prep_ligand,
properties=prop)
Visualizing small molecule (drug)
Visualizing the desired drug to be docked to the target protein, using NGL viewer.
Left panel: PDB-formatted file, with all hydrogen atoms.
Right panel: PDBqt-formatted file (AutoDock Vina-compatible), with united atom model (only polar hydrogens are placed in the structures to correctly type heavy atoms as hydrogen bond donors).
from ipywidgets import HBox
v0 = nglview.show_structure_file(ligand)
v1 = nglview.show_structure_file(prep_ligand)
v0._set_size('500px', '')
v1._set_size('500px', '')
def on_change(change):
v1._set_camera_orientation(change['new'])
v0.observe(on_change, ['_camera_orientation'])
HBox([v0, v1])

Preparing Target Protein for Docking
Preparing the target protein structure for the docking procedure. Converting the PDB file to a PDBqt file, needed by AutoDock Vina. Similarly to the previous step, the process adds partial charges and atom types to every target protein atom. In this case, however, we are not taking into account receptor flexibility, although Autodock Vina allows some limited flexibility of selected receptor side chains (see the documentation).
Building Blocks used:
str_check_add_hydrogens from biobb_structure_utils.utils.str_check_add_hydrogens
from biobb_structure_utils.utils.str_check_add_hydrogens import str_check_add_hydrogens
prep_receptor = "prep_receptor.pdbqt"
prop = {
"charges": True,
"mode": "auto"
}
str_check_add_hydrogens(
input_structure_path = pdb_protein,
output_structure_path = prep_receptor,
properties=prop)
Running the Docking
Running the docking process with the prepared files:
ligand
target protein
binding site box
using AutoDock Vina.
Building Blocks used:
autodock_vina_run from biobb_vs.vina.autodock_vina_run
from biobb_vs.vina.autodock_vina_run import autodock_vina_run
output_vina_pdbqt = "output_vina.pdbqt"
output_vina_log = "output_vina.log"
prop = { }
autodock_vina_run(input_ligand_pdbqt_path = prep_ligand,
input_receptor_pdbqt_path = prep_receptor,
input_box_path = output_box,
output_pdbqt_path = output_vina_pdbqt,
output_log_path = output_vina_log,
properties = prop)
Visualizing docking output poses
Visualizing the generated docking poses for the ligand, using NGL viewer.
Left panel: Docking poses displayed with atoms coloured by partial charges and licorice representation.
Right panel: Docking poses displayed with atoms coloured by element and ball-and-stick representation.
models = 'all'
v0 = nglview.show_structure_file(output_vina_pdbqt, default=False)
v0.add_representation(repr_type='licorice',
selection=models,
colorScheme= 'partialCharge')
v0.center()
v1 = nglview.show_structure_file(output_vina_pdbqt, default=False)
v1.add_representation(repr_type='ball+stick',
selection=models)
v1.center()
v0._set_size('500px', '')
v1._set_size('500px', '')
def on_change(change):
v1._set_camera_orientation(change['new'])
v0.observe(on_change, ['_camera_orientation'])
HBox([v0, v1])

Select Docking Pose
Select a specific docking pose from the output list for visual inspection.
Choose a docking pose from the DropDown list.
from Bio.PDB import PDBParser
parser = PDBParser(QUIET = True)
structure = parser.get_structure("protein", output_vina_pdbqt)
models = []
for i, m in enumerate(structure):
models.append(('model' + str(i), i))
mdsel = ipywidgets.Dropdown(
options=models,
description='Sel. model:',
disabled=False,
)
display(mdsel)
Extract a Docking Pose
Extract a specific docking pose from the docking outputs.
Building Blocks used:
extract_model_pdbqt from biobb_vs.utils.extract_model_pdbqt
from biobb_vs.utils.extract_model_pdbqt import extract_model_pdbqt
output_pdbqt_model = "output_model.pdbqt"
prop = {
"model": mdsel.value + 1
}
extract_model_pdbqt(input_pdbqt_path = output_vina_pdbqt,
output_pdbqt_path = output_pdbqt_model,
properties=prop)
Converting Ligand Pose to PDB format
Converting ligand pose to PDB format.
Building Blocks used:
babel_convert from biobb_chemistry.babelm.babel_convert
from biobb_chemistry.babelm.babel_convert import babel_convert
output_pdb_model = "output_model.pdb"
prop = {
"input_format": "pdbqt",
"output_format": "pdb",
"binary_path": "obabel"
}
babel_convert(input_path = output_pdbqt_model,
output_path = output_pdb_model,
properties=prop)
Superposing Ligand Pose to the Target Protein Structure
Superposing ligand pose to the target protein structure, in order to see the protein-ligand docking conformation.
Building a new PDB file with both target and ligand (binding pose) structures.
Building Blocks used:
cat_pdb from biobb_structure_utils.utils.cat_pdb
from biobb_structure_utils.utils.cat_pdb import cat_pdb
output_structure = "output_structure.pdb"
cat_pdb(#input_structure1 = pdb_single_chain,
input_structure1 = download_pdb,
input_structure2 = output_pdb_model,
output_structure_path = output_structure)
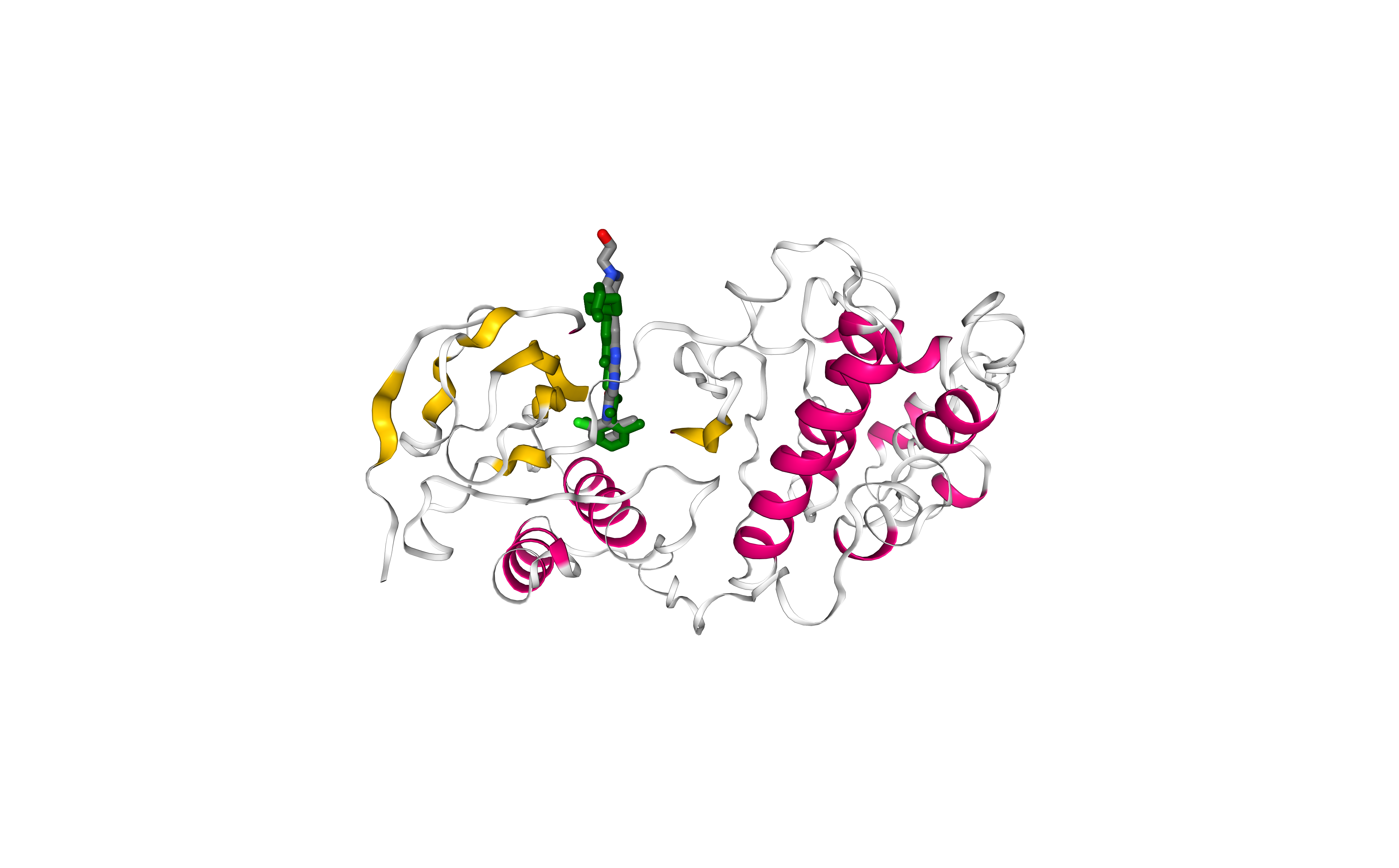
Comparing final result with experimental structure
Visualizing and comparing the generated protein-ligand complex with the original protein-ligand conformation (downloaded from the PDB database), using NGL viewer.
Licorice, element-colored representation: Experimental pose.
Licorice, green-colored representation: Docking pose.
Note that outputs from AutoDock Vina don’t contain all the atoms, as the program works with a united-atom representation (i.e. only polar hydrogens).
view = nglview.NGLWidget()
# v1 = Experimental Structure
v1 = view.add_component(download_pdb)
v1.clear()
v1.add_representation(repr_type='licorice',
selection='[1N1]',
radius=0.5)
# v2 = Docking result
v2 = view.add_component(output_structure)
v2.clear()
v2.add_representation(repr_type='cartoon', colorScheme = 'sstruc')
v2.add_representation(repr_type='licorice', radius=0.5, color= 'green', selection='UNL')
view._remote_call('setSize', target='Widget', args=['','600px'])
view
# align reference and output
code = """
var stage = this.stage;
var clist_len = stage.compList.length;
var i = 0;
var s = [];
for(i = 0; i <= clist_len; i++){
if(stage.compList[i] != undefined && stage.compList[i].structure != undefined) {
s.push(stage.compList[i])
}
}
NGL.superpose(s[0].structure, s[1].structure, true, ".CA")
s[ 0 ].updateRepresentations({ position: true })
s[ 0 ].autoView()
"""
view._execute_js_code(code)
view
Questions & Comments
Questions, issues, suggestions and comments are really welcome!
GitHub issues:
BioExcel forum: